
StudySmarter: Study help & AI tools
4.5 • +22k Ratings
More than 22 Million Downloads
Free
DNA is the basis of all living things on this planet. It is considered the code of life because it contains all the instructions that affect an organism's growth and development. It also carries the genetic information passed through the generations between parents and their offspring. The structure of DNA was explained back in 1953 by Rosalind Franklin, Maurice Wilkins, James Watson and Francis Crick. However, it would take many years before these fragments of DNA can be thoroughly analyzed by scientists. Today, DNA sequencing is a routine among scientists to understand and decode the genetic information present within the DNA.


Lerne mit deinen Freunden und bleibe auf dem richtigen Kurs mit deinen persönlichen Lernstatistiken
Jetzt kostenlos anmeldenDNA is the basis of all living things on this planet. It is considered the code of life because it contains all the instructions that affect an organism's growth and development. It also carries the genetic information passed through the generations between parents and their offspring. The structure of DNA was explained back in 1953 by Rosalind Franklin, Maurice Wilkins, James Watson and Francis Crick. However, it would take many years before these fragments of DNA can be thoroughly analyzed by scientists. Today, DNA sequencing is a routine among scientists to understand and decode the genetic information present within the DNA.
DNA sequencing is the process of determining the DNA nucleotide sequence, or the order of bases that make up a DNA segment. We can use this information to determine the RNA or protein sequence that leads to more information about the gene’s function and its relationship to other genes. We can also use this information to study gene expression and regulation. To understand DNA sequencing, we must first understand the structure of DNA.
DNA has a double helix structure composed of building blocks we call nucleotides (or bases). DNA is composed of four building block nucleotides. These bases are divided into two categories namely purine bases which are Guanine (G) and Adenine (A) and pyrimidine bases which are Cytosine (C) and Thymine (T). A strand of DNA would be composed of A, G, C, and T, repeating in a seemingly random order (Fig. 1).
At first, the order of these four bases may seem random, but it is not random at all. The arrangement of these four bases is very important and corresponds to different genetic information within a cell or an organism. These bases provide the underlying genetic basis for different traits in an individual. (also known as his phenotype)
Let's say the DNA sequence CGATGG transmits genetic information for black hair. Even if there is only a difference of one base, the DNA sequence CGATCG might transmit genetic information for brown hair.
This genetic information is crucial in understanding the basis of genetic diseases like Huntington’s disease, cystic fibrosis, Down syndrome, and many others. Knowing a DNA sequence is key to understanding the function of our genes.
Any change in this DNA sequence is called a mutation. You can think of mutation as a "mistake" in the DNA sequence that can arise when the DNA is copied during DNA replication or as a result of different environmental factors such as smoking, exposure to sunlight, radiation, and other mutagens.
Mutation in DNA can lead to diversity in species as it produces new alleles (gene variants). Mutations may be harmful, beneficial, or neutral. Harmful mutations negatively impact an organism's evolutionary fitness or ability to survive and reproduce. On the contrary, beneficial mutations positively impact an organism's evolutionary fitness. Most mutations are neutral: they have no effect on an organism’s evolutionary fitness. While most mutations are neutral, more serious mutations can lead to various lethal genetic disorders. One of the most common human genetic diseases is cancer caused by harmful mutations, leading to the uncontrolled growth of cells.
The four nitrogenous bases pair up and are joined by hydrogen bonds. Adenine (A) always pairs with thymine (T), joined by two hydrogen bonds, while Cytosine (C) always pairs with guanine (G), joined by three hydrogen bonds. This is called complementary base pairing. Complementary base pairing plays an important role in DNA sequencing.
Gene expression is the process of converting instructions in our DNA into RNA and protein. It takes place in two major stages: transcription, where a copy of a gene's DNA sequence is produced and written into RNA, and translation, where protein is synthesized using the genetic information contained in the messenger RNA (mRNA) template. Let's examine how a DNA sequence transforms during these two stages of gene expression.
During DNA transcription, the DNA strand serves as a template for mRNA. RNA polymerase enzyme forms an mRNA by travelling through the DNA strand from 3′ → 5’ end. As it travels through the strand, it “copies” the sequence of the bases by adding complementary base pairs from 5′ → 3′ end. Recall that RNA has uracil (U) instead of thymine (T) while retaining adenine (A), guanine (G), and cytosine (C). A guanine (G) in DNA would indicate the addition of a cytosine (C) into the growing mRNA strand. Similarly, a thymine (T) in DNA will be copied into an adenine (A) in the mRNA. The information in the DNA sequence would be passed onto this mRNA. The mRNA will then undergo translation to produce a protein.
The mRNA moves from the nucleus (in eukaryotes) or the cytoplasm (in prokaryotes) to the ribosomes, where it will be translated into proteins. The order of the mRNA bases would correspond to specific amino acids, which are the building blocks of proteins.
As mentioned earlier, DNA contains the genetic information needed to produce proteins. The DNA sequence transcribed into mRNA will then be used to form amino acid chains, which make up protein. Every three bases in an mRNA would correspond to one codon, and each codon specifies an amino acid (Fig. 2).
Sequencing DNA requires breaking apart the DNA sequence into smaller chunks or fragments. The order of bases of these small fragments is determined and then assembled to make up the original fragment.
One of the most popular techniques for DNA sequencing is the Sanger sequencing method or the chain termination method. It is considered a “first-generation” sequencing method. In Sanger sequencing, the DNA sequence of interest is amplified like that of a polymerase chain reaction but modified in such a way that it is now a chain termination polymerase chain reaction.
A polymerase chain reaction is a laboratory technique in which a DNA segment is "amplified", meaning millions to billions of copies of the segment are created. This process uses primers (short synthetic DNA fragments) to determine which segment will be amplified. DNA synthesis is then done several times to amplify that segment.
Chain Termination polymerase chain reaction follows a conventional polymerase chain reaction, except it contains an additional modified dideoxy or chain-terminating nucleotides called deoxyribonucleotides (ddNTPs) which are also uniquely fluorescently labelled.
First, the double-stranded DNA would be denatured through heating. Once cooled, a primer would be attached to the single-stranded DNA template. Upon raising the temperature again, the extension step will begin: a DNA polymerase adds nucleotides to synthesize new DNA until it adds a ddNTP from the mixture, which terminates the whole reaction.
This cycle will be repeated multiple times, ensuring that a ddNTP is virtually added at every position of the DNA sequence. This will result in multiple fragments of DNA of varying lengths. Since the end of the fragments is fluorescently labelled, this would indicate the final nucleotide that was added. The mixture of fragments will be run through capillary gel electrophoresis, which separates the fragments through size.
A detector will be able to detect the fluorescent signals resulting in a chromatogram. A chromatogram typically shows the results of DNA sequencing, where the four bases are represented by specific colors (Fig. 3). A chromatogram usually contains 1000 to 1200 bases.
The DNA sequence can now be determined through such chromatogram by reading the bases from left to right. This order is equivalent to the sequence of bases from the 5' to 3' end of the DNA strand. While Sanger sequencing is effective in sequencing small fragments of DNA, even up to 900 base pairs, sequencing larger fragments would be highly inefficient using this technique. For large-scale sequencing, like sequencing an organism's genome, recent DNA sequencing technologies called next-generation sequencing are used.
Improvements in DNA sequencing led to the development of newer second-generation or next-generation sequencing (NGS). The principle behind NGS is similar to that of Sanger sequencing. NGS involves three general steps:
Library preparation
DNA Amplification
DNA Sequencing
During library preparation, the starting DNA is cut into random fragments either mechanically or enzymatically.
Once a library of different fragment sizes of DNA is produced, it will be amplified through a polymerase chain reaction.
Once a suitable library is prepared, DNA needs to be amplified in order for a sequencer to detect the signal. During amplification, a primer would bind to the single-stranded template by complementary base pairing. This primer will be the starting point for a Taq polymerase to add bases and make new strands of DNA.
Because of the complementary base pairing of DNA, researchers can predict the sequence of the complementary DNA once the sequence of a DNA strand is known. This complementary base pairing is the basis of the Taq polymerase in synthesizing new strands of DNA.
Sequencing is done using various NGS methods (these include Illumina, pyrosequencing, and sequencing by ligation). This is done by loading the library onto the sequencing platform, which reads the bases and produces data that will be analyzed by specialized software.
Before, sequencing an entire genome would be unthinkable. Scientists then did not have the tools and techniques to analyze large DNA fragments.
Today, with the advent of new technologies, scientists are able to sequence whole genomes of different organisms as shown in the Human Genome Project which lasted from 1990 to 2003. The Human Genome Project was a collaboration among a team of international scientists which aimed to completely sequence the whole human genome and map the location of important genes in our chromosomes.
They were able not just to determine the base pairs of DNA but also to map all of the genes and annotate some of its function. This endeavor has led to very important discoveries about the structure, organization, and function of the human genome.
An unintended benefit of the project was the development of faster and cheaper methods of DNA sequencing. In 2001, sequencing 1 million bases would cost over $5,000. This has decreased to $0.02 in 2016. Furthermore, whereas sequencing the first human genome took over 10 years, sequencing a human genome today would take just a couple of days.
DNA sequencing is the process of determining the DNA sequence, or the order of bases that make up a DNA segment.
DNA can be cut enzymatically using restriction enzymes (RE). After recognizing sequence-specific sites, REs cleave DNA by producing a blunt or sticky end with a known sequence at each end.
A change in a cell's dna sequence is mutation.
DNA sequencing works by breaking apart the DNA sequence into smaller chunks or fragments.
A DNA sequence is transcribed by an enzyme called RNA polymerase which travels through the strand and "copies” the sequence of the bases by adding complementary base pairs from 5′ → 3′.
What is DNA sequencing?
DNA sequencing is the process of determining the arrangement of nucleotide bases in a DNA segment.
Any change in a DNA sequence is called
Mutation
How does a DNA sequence form protein?
A DNA sequence is transcribed into mRNA, which is then translated into protein.
A diagram that shows the results of DNA sequencing, where the four bases are represented by specific colors is called ___.
Chromatogram
The Sanger method is considered a __.
first-generation method of sequencing
__ is a laboratory technique in which a DNA segment is "amplified", meaning millions to billions of copies of the segment are created.
Polymerase chain reaction
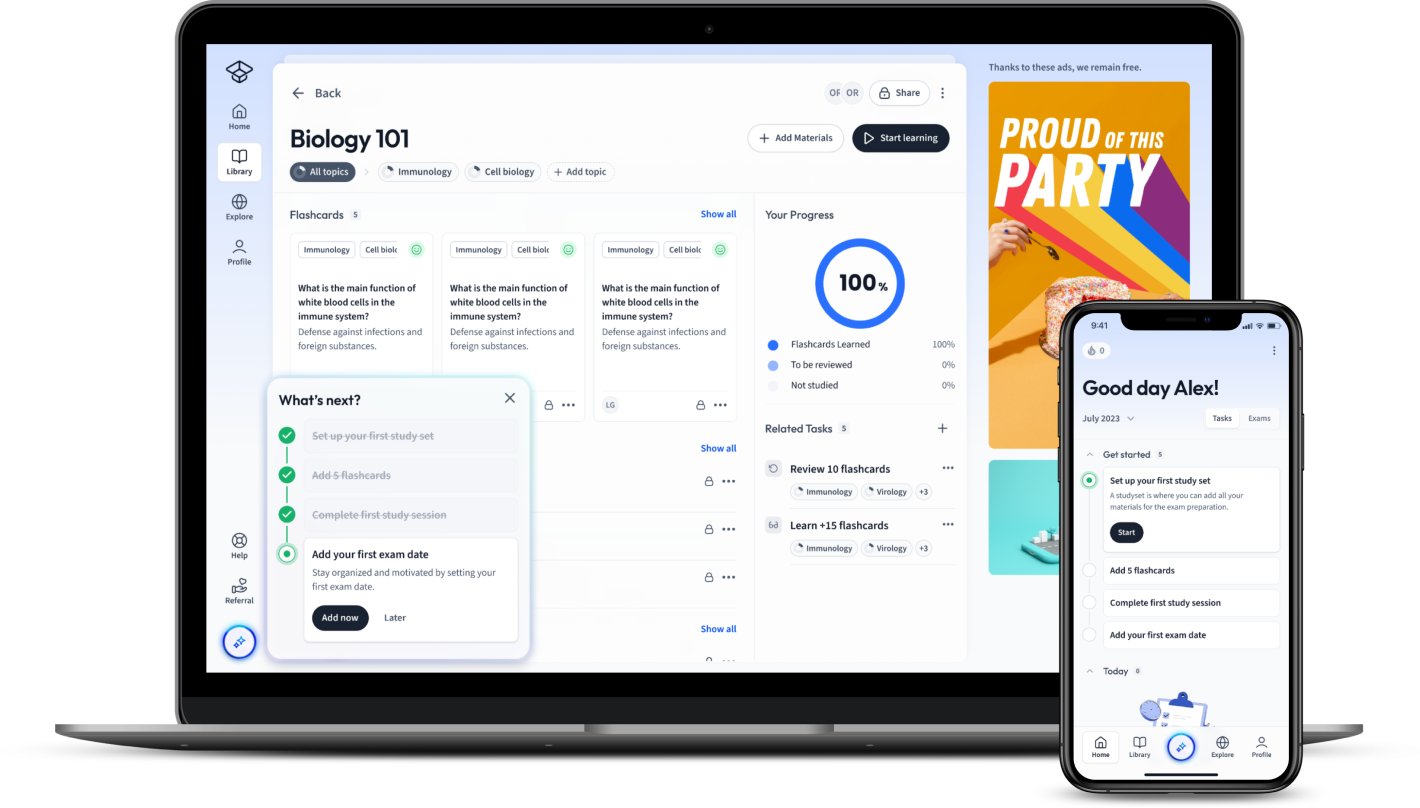
Already have an account? Log in
Open in AppThe first learning app that truly has everything you need to ace your exams in one place
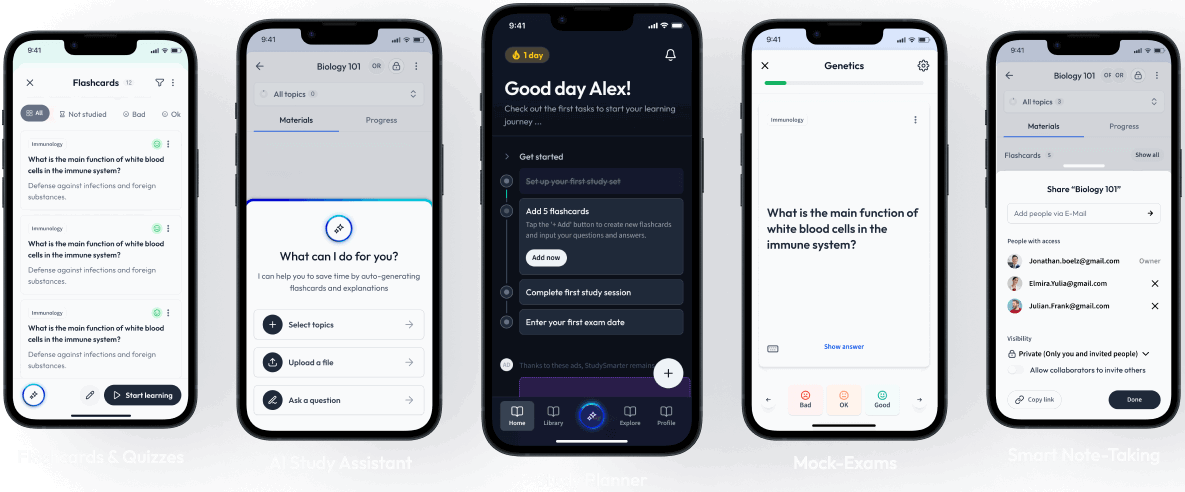
Sign up to highlight and take notes. It’s 100% free.
Save explanations to your personalised space and access them anytime, anywhere!
Sign up with Email Sign up with AppleBy signing up, you agree to the Terms and Conditions and the Privacy Policy of StudySmarter.
Already have an account? Log in